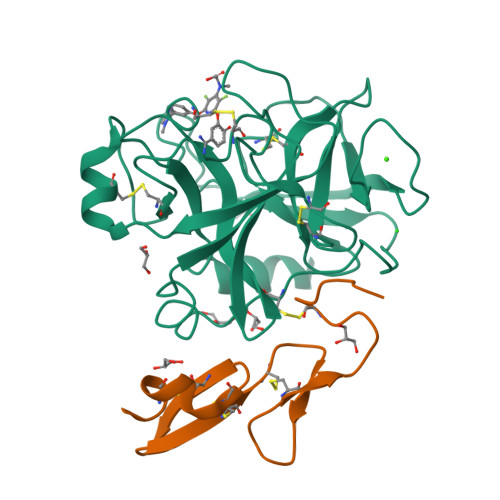
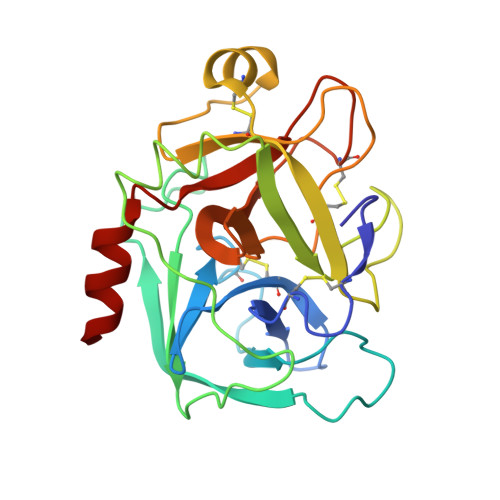
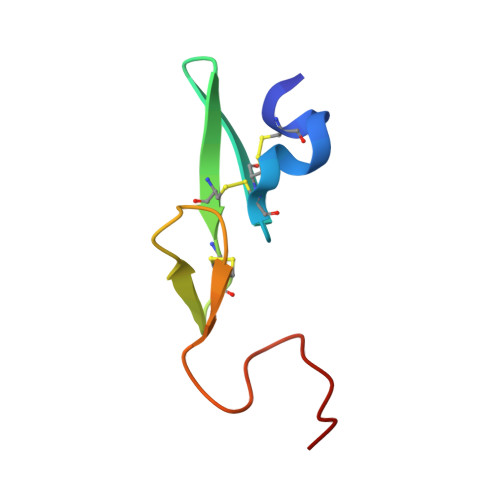
Preparation, characterization, and the crystal structure of the inhibitor ZK-807834 (CI-1031) complexed with factor Xa.
Adler, M., Davey, D.D., Phillips, G.B., Kim, S.H., Jancarik, J., Rumennik, G., Light, D.R., Whitlow, M.(2000) Biochemistry 39: 12534-12542
- PubMed: 11027132
- DOI: https://doi.org/10.1021/bi001477q
- Primary Citation of Related Structures:
1FJS - PubMed Abstract:
Factor Xa plays a critical role in the formation of blood clots. This serine protease catalyzes the conversion of prothrombin to thrombin, the first joint step that links the intrinsic and extrinsic coagulation pathways. There is considerable interest in the development of factor Xa inhibitors for the intervention in thrombic diseases. This paper presents the structure of the inhibitor ZK-807834, also known as CI-1031, bound to factor Xa and provides the details of the protein purification and crystallization. Results from mass spectrometry indicate that the factor Xa underwent autolysis during crystallization and the first EGF-like domain was cleaved from the protein. The crystal structure of the complex shows that the amidine of ZK-807834 forms a salt bridge with Asp189 in the S1 pocket and the basic imidazoline fits snugly into the S4 site. The central pyridine ring provides a fairly rigid linker between these groups. This rigidity helps minimize entropic losses during binding. In addition, the structure reveals new interactions that were not found in the previous factor Xa/inhibitor complexes. ZK-807834 forms a strong hydrogen bond between an ionized 2-hydroxy group and Ser195 of factor Xa. There is also an aromatic ring-stacking interaction between the inhibitor and Trp215 in the S4 pocket. These interactions contribute to both the potency of this compound (K(I) = 0.11 nM) and the >2500-fold selectivity against homologous serine proteases such as trypsin.
Organizational Affiliation:
Berlex Biosciences, 15049 San Pablo Avenue, P.O. Box 4099, Richmond, California 94804-0099, USA.